Biography
"My long-term motivation is to become an interdisciplinary, system-scale thinker aiming to achieve a macro-level understanding of biological diversity."
I am postdoctoral researcher at the Molecular Biology Department, Max Planck Institute for Biology. I am part of the Geogenomic Archaeology Campus of Tübingen (GACT) project, mainly involved in the Biomolecules & Bioinformatics Division. The main goal of GACT is to measure the impact of humans in cave ecosystems through metagenomic characterisation of sediments and coprolites ancient DNA (aDNA). I am very happy and feel very fortunate to announce that during this new stage I will be surrounded by a big group of amazing people and scientists driven by curiosity in the DrostLab, Weigel World and Archaeo- and Paleogenetics group supervised by Hajk-Georg Drost, Detlef Weigel and Cosimo Posth, respectively.
I have done my PhD with EPIOMIDES group, lead by María Jesús Cañal, Mónica Meijón and Luis Valledor, in the Organisms and Systems Biology Department at the University of Oviedo. My thesis focused on abiotic stress responses in different species, specifically using unexplored gymnosperms as models, from a Systems Biology approach.
Beyond my professional responsabilities i am also a being half passionate half obssesed with bioinformatics and computational biology. When i have free time (wait for it xd), really enjoy trying new softwares specially those ones related with evolutionary genomics and transcriptomics.
When i am not in the lab or the batcave (home office), I dance breaking (Break Dance) and try not to become an e-boy while exchanging with other art forms.
I will add more info in the future of upcoming projects!
Publications
Proteomic dynamics revealed sex‐biased responses to combined heat‐drought stress in Marchantia
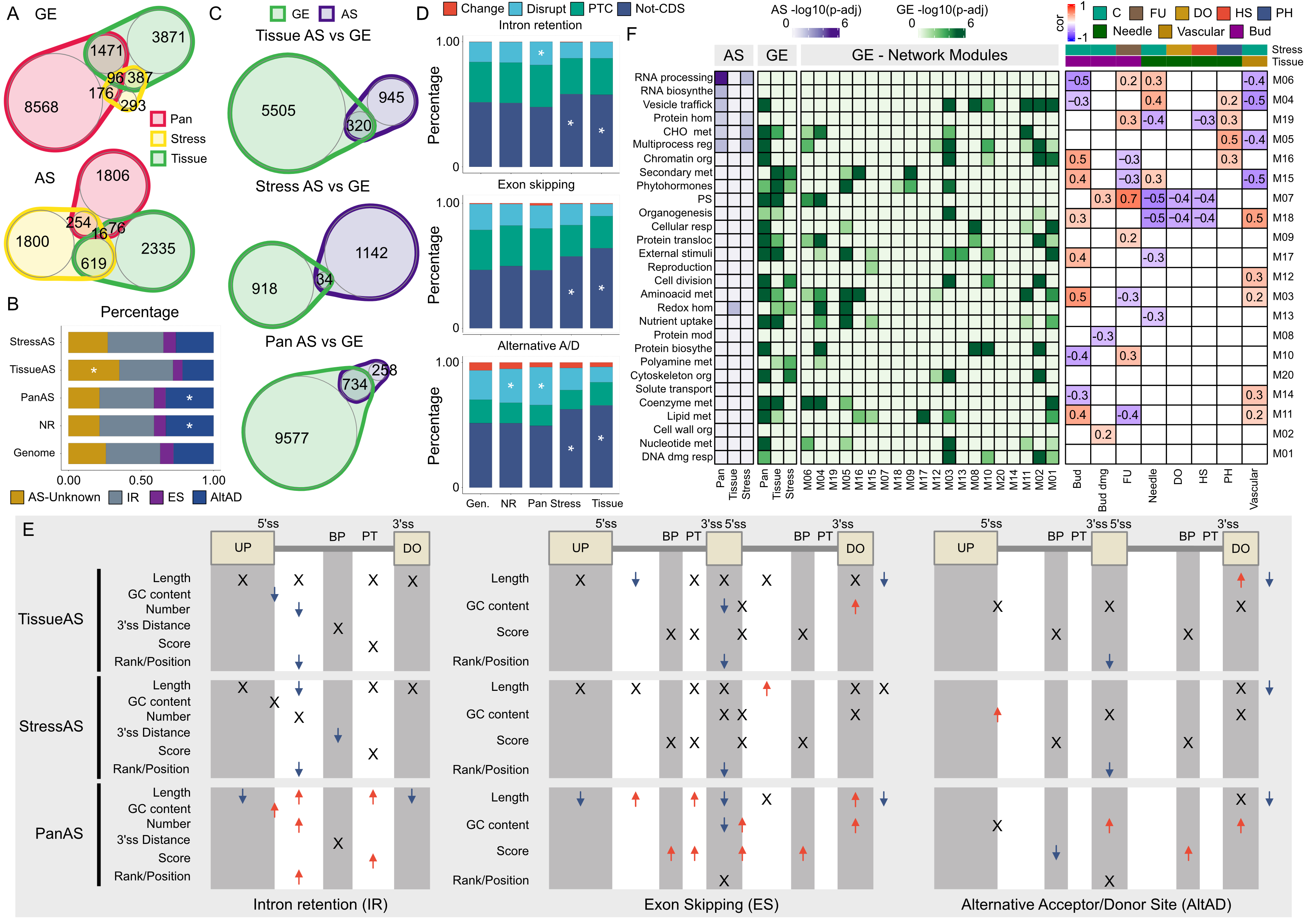
Pra-GE-ATLAS: empowering Pinus radiata stress and breeding research through a comprehensive multi-omics database
PlantFUNCO: integrative functional genomics database reveals clues into duplicates divergence evolution

Long term splicing-memory and its divergence in Spermatophyta revealed by integrative heat dynamics in Pinus
Protein Interaction Networks (PPIs): Functional and Statistical Approaches
Tools
Chlamytina: displaying Chlamydomonas reinhardtii epipotreomic landscape
Chlamytina is a project focused in this well-known green-alage model try to answer a common question in some proteomic/transcriptomic studies: Are my molecules of interest epigenetically regulated?
To fill this gap, we collected all epigenectic files published until the date and developed a new chromatin states model (ChromHMM) including 6mA, 5mC and nucleosome-profile for the first time. Additionally, an epigenome-browser was conducted focusing on the site-specific approach. This tool engage the link-up between proteomic/transcriptomic changes and epigenetic patterns, thus displaying the Chlamydomonas reinhardtii epi-proteogenomic landscape.
P(inus)ra(diata)-G(ene)E(expression) Atlas
In the current context of global warming, the study of stress in plants becomes a priority. Conifers have received little attention even though their phylogenetic position can solve several key evolutionary and environmental acclimation questions. Pinus, the largest genus of conifers and arguably the most important genus of trees in the world, provides an ideal example to explore gene regulation differentiation due to its long evolutionary history and potentially unique genomic features. We focused in Pinus radiata the most stress-sensitive species in the genus and widely used for contanst production of forest services. Pra-GE Atlas tries to answer the next questions: Where and when are all genes identified till the date expressed? , Which is the regulatory signals defining tissue identity? Specific and conserved responses to environmental stress?
This is a challenge because P.radiata as a non-model organism does not have public reference genome, despite its industrial interest. This tool will generate a large amount of basic knowledge about this organism and will make applied research easier in the future.
PlantFUNCO: integrative functional-genomics to allow the detection of functional constraints
Many comparative-genomics studies interrogate sequence-conserved loci of interest across a wide range of species and its function is determined by perturbing their homologous in a single model organism. In this context, a maze of opportunities and challenges appeared to systematically and confidently determine the extent of conservation at the functional genomics level between model species. PlantFUNCO is constituted by various tools and two main resources, inter-species chromatin states and functional genomics conservation scores, for three well known plant model organisms (A.thaliana, Z.mays and O.sativa).
PlantFUNCO aim to leverage data diversity and extrapolate findings from different models to determine the extent of functional constraint, thus, deepen our understanding of how plants phenotypic plasticity has fascinatingly evolved.
EDUCATION
CD, Biology, University of Oviedo, Spain (2014-2018)
MS, Plant Biotechnology, University of Oviedo, Spain (2018-2019)
Short PhD Stay, Max Planck Institute - Biology, Computation Biology Group DrostLab-WeigelWorld, Tübingen, Germany (2023)
PhD, Biology, University of Oviedo, Spain (2019-2024)
Postdoc, Bioinformatics, Max Planck for Biology - GACT, Germany (2024-now)
GRANTS
Beca de colaboración, Min. de Educación y Formación Profesional ,2016
Formación de profesorado universitario, Mi. de Ciencia, Innovación y Universidades, 2018
Mobility grant: Formación de profesorado universitario, Mi. de Ciencia, Innovación y Universidades, 2023